We study how transcription factors bind to the correct DNA sequences and regulate the correct target genes in vivo, with a focus on the Hox family of homeodomain proteins.
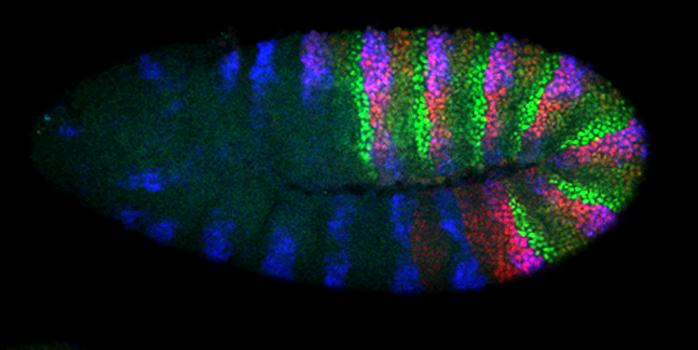
The Hox genes encode a conserved family of homeodomain-containing transcription factors that specify tissue and cellular identities throughout the animal kingdom. As is typical for homeodomain proteins, they tend to bind a wide range of degenerate, TAAT-containing binding sites, raising the fundamental question of how these factors achieve specificity in vivo. We use a variety of approaches, including whole genome binding studies, in vitro DNA binding assays, X-ray crystallography, in vivo functional tests, and high-throughput DNA specificity measurements, to address this question. Some of our work (Slattery et al., 2011) reveals that Hox cofactors reveal “latent specificity” that is present in the Hox homeodomain, but cannot be used in the absence of the cofactors. In one specific case (Scr; Joshi et al., 2007) we found that latent specificity allows normally unstructured residues in the Hox protein Scr to read a DNA shape: the width of the minor groove. We are building upon these results by leveraging whole genome experiments and classical Drosophila genetics to gain a deeper understanding for how transcription factors orchestrate developmental processes.
Current lab members working on this project: Bill Glassford, Siqian Feng, Ryan Loker, Ross Munce, and Rebecca Delker (collaborations with Barry Honig, Remo Rohs and Harmen Bussemaker).
Publications
Dense and pleiotropic regulatory information in a developmental enhancer. Fuqua T, Jordan J, van Breugel ME, Halavatyi A, Tischer C, Polidoro P, Abe N, Tsai A, Mann RS, Stern DL, Crocker J. Nature. 2020 Nov;587(7833):235-239. doi: 10.1038/s41586-020-2816-5. Epub 2020 Oct 14. PMID: 33057197.
Context-Dependent Gene Regulation by Homeodomain Transcription Factor Complexes Revealed by Shape-Readout Deficient Proteins. Kribelbauer JF, Loker RE, Feng S, Rastogi C, Abe N, Rube HT, Bussemaker HJ, Mann RS. Mol Cell. 2020 Apr 2;78(1):152-167.e11. doi: 10.1016/j.molcel.2020.01.027. Epub 2020 Feb 12. PMID: 32053778
Toward a Mechanistic Understanding of DNA Methylation Readout by Transcription Factors. Kribelbauer JF, Lu XJ, Rohs R, Mann RS, Bussemaker HJ. J Mol Biol. 2019 Nov 2:S0022-2836(19)30617-5. doi: 10.1016/j.jmb.2019.10.021. PMID: 31689433
Low affinity binding sites in an activating CRM mediate negative autoregulation of the Drosophila Hox gene Ultrabithorax. Delker RK, Ranade V, Loker R, Voutev R, Mann RS. PLoS Genet. 2019 Oct 7;15(10):e1008444. doi: 10.1371/journal.pgen.1008444. eCollection 2019 Oct. PMID: 31589607
In vivo Hox binding specificity revealed by systematic changes to a single cis regulatory module. Sánchez-Higueras C, Rastogi C, Voutev R, Bussemaker HJ, Mann RS, Hombría JC. Nat Commun. 2019 Aug 9;10(1):3597. doi: 10.1038/s41467-019-11416-1. PMID: 31399572
Low-Affinity Binding Sites and the Transcription Factor Specificity Paradox in Eukaryotes. Kribelbauer JF, Rastogi C, Bussemaker HJ, Mann RS. Annu Rev Cell Dev Biol. 2019 Oct 6;35:357-379. doi: 10.1146/annurev-cellbio-100617-062719. Epub 2019 Jul 5. PMID: 31283382
Tim Zeiske, Nithya Baburajendran, Anna Kaczynska, Julia Brasch, Arthur G. Palmer III, Lawrence Shapiro, Barry Honig, Richard S. Mann Intrinsic DNA shape accounts for affinity differences between Hox- cofactor binding sites. Cell Reports, Volume 24, Issue 9, 28 August 2018, Pages 2221-2230. August 2018
Chaitanya Rastogi, H. Tomas Rube, Judith F. Kribelbauer, Justin Crocker, Ryan E. Loker, Gabriella D. Martini, Oleg Laptenko, William A. Freed-Pastor, Carol Prives, David L. Stern, Richard S. Mann and Harmen J. Bussemaker Accurate and sensitive quantification of protein-DNA binding affinity. Proceedings of the National Academy of Sciences Apr 2018, 201714376; DOI: 10.1073/pnas.1714376115. April 2018
Judith Kribelbauer, Oleg Laptenko, Siying Chen, Gabriella Martii, William Freed-Pastor, Carol Prives, Richard S. Mann, Harmen Bussemaker Quantitative Analysis of the DNA Methylation Sensitivity of Transcription Factor Complexes Cell Reports. 19, 2383–2395. June 2017
Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, Mann RS, Stern DL. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell. 2015 Jan 15;160(1-2):191-203. doi: 10.1016/j.cell.2014.11.041. Epub 2014 Dec 31. PubMed PMID: 25557079; PubMed Central PMCID: PMC4449256.
Abe N, Dror I, Yang L, Slattery M, Zhou T, Bussemaker HJ, Rohs R, Mann RS. Deconvolving the recognition of DNA shape from sequence. Cell. 2015 Apr 9;161(2):307-18. doi: 10.1016/j.cell.2015.02.008. Epub 2015 Apr 2. PubMed PMID: 25843630; PubMed Central PMCID: PMC4422406.
Merabet S, Mann RS. To Be Specific or Not: The Critical Relationship Between Hox And TALE Proteins. Trends Genet. 2016 Jun;32(6):334-47. doi: 10.1016/j.tig.2016.03.004. Epub 2016 Apr 8. Review. PubMed PMID: 27066866; PubMed Central PMCID: PMC4875764.
